
|
Synthèse de Molécules et de Macromoléculespour le Vivant et l’Environnement |
Chemical tools for the study of epigenetic modifications
Epigenetics defines the changes in gene expression that are not coded by the DNA sequence and are inheritable. The main epigenetic factors are chemical modification of histones (methylation and acetylation) and DNA methylation, but new modifications have recently expanded the epigenetic landscape. Considering the potential of “druggability” of proteins involved in epigenetics, the precise characterization of their activity might offer new opportunity for therapeutic intervention. In recent years, the study of chromatin biology has greatly benefited from chemical biology tools to address fundamental structure/function problems in epigenetics. We recently developed both chemical tools and proteomic approaches based on photolabeling or bioorthogonal labeling coupled to mass spectrometry to study several epigenetic modifications.
Identification of Novel Inhibitors of DNA methylation by Screening of a Chemical Library. ACS Chem Biol. 2013, 8 (3), pp 543–548.
Synthesis and Evaluation of Analogues of N-Phthaloyl-L-tryptophan (RG108) as Inhibitors of DNA Methyltransferase 1. Journal of Medicinal Chemistry. 2014, 57(2), 421-34.
Combined analysis of DNA methylation cell cycle in cancer celles. Epigenetics. 2015, 10(1), 82-91.
A direct label-free MALDI-TOF mass spectrometry based assay for the characterization of inhibitors of protein lysine methyltransferases. Analytical and Bioanalytical Chemistry. 2017, 409(15), 3767–3777.
Rational design of bisubstrate-type analogs as inhibitors of DNA methyltransferases in cancer cells. Journal of Medicinal Chemistry. 2017, 60 (11), 4665–4679.
Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors. Philos Trans R Soc Lond B Biol Sci. 2018, 5, 373(1748)
Photoactivatable oligonucleotide probes to trap single-stranded DNA binding proteins: Updating the potential of 4-thiothymidine from a comparative study. Biochimie. 2018, 154, 164-175.
Trehalose and mycolic acid-based probes for the study of mycomembrane biogenesis
Recently, chemical-biology-based strategies have emerged for the study of bacterial pathogens, giving biologists the opportunity to have powerful new tools for probing bacteria. Actually, probes for the study of mycobacterial envelope remains challenging since Mycobacteria and Corynebacteria have an atypical organization being composed of complex glycolipids (mycolic acids esterified to α,α-D-trehalose). They form a very impermeable and rigid barrier that might contribute to the exceptional resistance of mycobacteria towards chemotherapeutic molecules. The biosynthesis of these cell wall mycolate esters has been studied during the last decade but the mechanisms of their assembly in the bacterial envelope is largely unknown.
In this context, we study the specificity and the interplay between several enzymes named mycoloytransferases in the biogenesis of the envelope of these mycobacteria, using our expertise in trehalose chemistry including the synthesis of complex fatty acids and selective modifications of trehalose. We work in this project in close collaboration with Prof. Nicolas Bayan (Institut de Biologie Intégrative de la Cellule, Université Paris Sud), Dr. Boris Vauzeilles (Chemical Biology Department, Institut de Chimie des Substances Naturelles) and with Prof. Federica Migliardo (University of Messina).
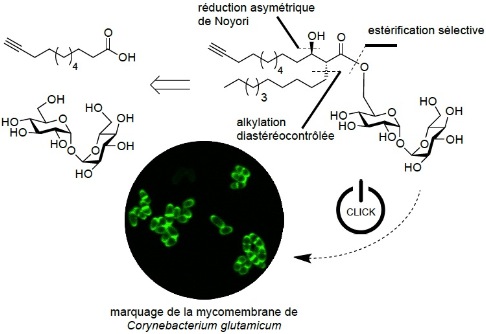
Synthesis of chemical tools to label the mycomembrane of corynebacteria using a modified Iron (III) chloride-mediated protection of trehalose. M. Carlier, E. Lesur, A. Baron, A. Lemétais, K. Guitot, L. Roupnel, C. Dietrich, G. Doisneau, D. Urban, N. Bayan, J.-M. Beau, D. Guianvarc’H, B. Vauzeilles, Y. Bourdreux, Org. Biomol. Chem., 2022, 20, 1974-1981
Etudier la membrane des mycobactéries via une approche chémobiologique. E. Lesur, Y. Bourdreux, L'Actualité Chimique, 2021, 465, 63-64
Dominique Guianvarc'h, Yann Bourdreux, Christophe Biot, Boris Vauzeilles, Metabolic Labeling of Bacterial Glycans, Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, Elsevier, 2021, ISBN 9780124095472, https://doi.org/10.1016/B978-0-12-819475-1.00098-5
"First access to a mycolic acid-based bioorthogonal reporter for the study of the mycomembrane and mycoloyltransferases in Corynebacteria". E. Lesur, A. Baron, C. Dietrich, M. Buchotte, G. Doisneau, D. Urban, J.-M. Beau, N. Bayan, B. Vauzeilles, D. Guianvarc’h, Y. Bourdreux, Chem. Commun., 2019, 55, 13074-13077.
"Study of the conformational behaviour of trehalose mycolates by FT-IR spectroscopy". F. Migliardo, Y. Bourdreux, M. Buchotte, G. Doisneau, J.-M. Beau, N. Bayan, Chem. Phys. Lipids, 2019, 223, 104789
For the selective synthesis of Mycobacterium tuberculosis sulfoglycolipids, see:
"Synthesis of a Mycobacterium tuberculosis Tetra-acylated Sulfolipid Analogue and Characterization of the Chiral Acyl Chains Using Anisotropic NAD 2D-NMR Spectroscopy"" A. Lemétais, Y. Bourdreux, P. Lesot, J. Farjon, J.-M. Beau, J. Org. Chem. 2013, 78, 7648–7657.
"Simplified Deoxypropionate Acyl Chains for Mycobacterium tuberculosis Sulfoglycolipid Analogues: Chain Length is Essential for High Antigenicity" B. Gau, A. Lemétais, M. Lepore, L. F. Garcia-Alles, Y. Bourdreux, L. Mori, M. Gilleron, G. De Libero, G. Puzo, J.-M. Beau, J. Prandi, ChemBioChem 2013, 14, 2413–2417.
"Recent results in synthetic glycochemistry with iron salts at Orsay-Gif" J.-M. Beau, Y. Bourdreux, F.-D. Boyer, S. Norsikian, D. Urban, G. Doisneau, B. Vauzeilles, A. Gouasmat, A. Lemétais, A. Mathieu, J.-F. Soul., A. Stévenin, A. Xolin, in Carbohydrate Chemistry, vol. 40 (Eds.: A. P. Rauter, T. K. Lindhorst, Y. Queneau), The Royal Society of Chemistry, Cambridge, UK, 2014, pp. 118–139.

