CP3A ORGANIC SYNTHESIS GROUP
CP3A ORGANIC SYNTHESIS GROUP
Peptidomimetic Chemistry, Photochemistry, Alternative Processes
Bottom-up design features for peptide-based foldamers
Short oligo-peptides constructed from non-canonical and often homologated amino acids have been a cornerstone of foldamer chemistry since its inception, and they remain the focus of a considerable part of contemporary research. Much remains to be discovered, and the continued expansion of the monomer toolbox should an asset for further explorations. Our group has focused on the particular interest of foldamer manifolds which feature homologated amino acids bearing a four-membered ring. For our synthetic work providing a small library of such building blocks, see this page. We study the folding information which is embedded in these monomers, then we endeavor to apply this information in the bottom-up design of foldamers with predictable architectures.
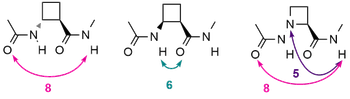
We have conducted detailed studies on cis and trans 2-cyclobutane β-aminoacids (ACBC), as well as the aza-analogue (AAzC). The local folding preferences of these monomers are a strong 8-membered ring for trans-ACBC and a weaker 6-membered ring for cis-ACBC; AAzC behaves as an aza-analogue of trans-ACBC and has a strong propensity for the hydrazino turn, which strengthens the 8-membered ring hydrogen bond. Chem. Eur. J., 2015, 21, 16479-16493; J. Org. Chem., 2013, 78, 6031-6039; Tetrahedron Lett., 2013, 54, 802-805.
|
|
Hydrazino turn: A bifurcated hydrogen bond implicating the sp3 nitrogen |
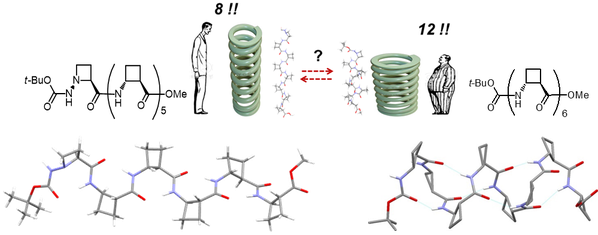
Our group was the first to show that, despite the capacity of trans-ACBC to support a short-range 8-membered ring, homo-oligomers with more than four residues adopt a regular 12-helix structure, articulated around a repeating series of N-H(i)…O=C(i–3) hydrogen bonds. Using the hydrazino turn feature of AAzC as a conceived 8-helix primer at the N-terminal, we found that a short trans-ACBC sequence was willing to follow the lead, and adopt an alternative 8-helix conformer up to a chain length of six residues. Angew. Chem. Int. Ed., 2015, 54, 10807-10810; Org. Lett., 2010, 12, 3606-3609.
Using trans-ACBC as a programmed helix-inducer, we devised mixed β,γ-peptides which adopt 13-helix conformations, relying on minimal steric imposition. The topology of these foldamers is particularly interesting: they give excellent super-position upon a model of a native α-helical peptide backbone. Chem. Commun., 2016, 52, 7802-7805.
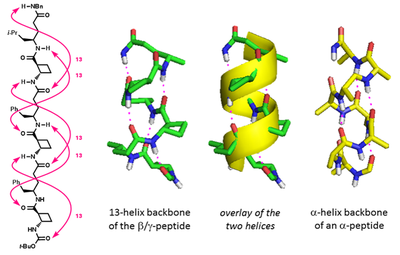
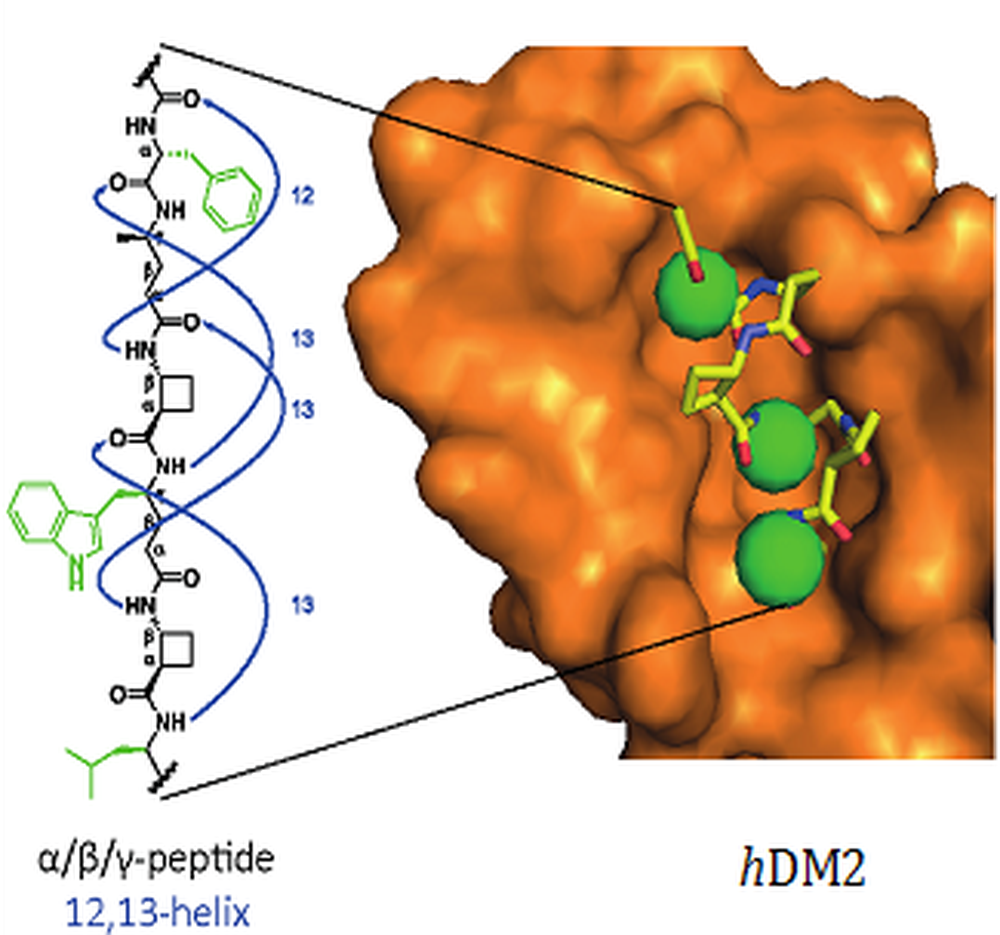
We have used this discovery as the central feature of a bottom-up foldamer design rational for functional mimics of protein secondary structures. In collaboration with Andy Wilson (Leeds) we prepared a series of α,β,γ-peptides which adopt a robust 12,13-helix conformation in solution and act as selective inhibitors of the p53/hDM2 protein–protein interaction. Angew. Chem. Int. Ed., 2016, 55, 11096-11100.
|
A helical mimetic of the [Phe19-Leu26] segment of the p53 protein. This segment bears three "hot-spot" residues implicated in its interaction with the hDM2 protein. The hybrid α,β,γ-peptide inhibits the p53/hDM2 interaction with IC50 = 15 mM (competition assay using fluorescence anisotropy) by binding to the natural binding site of the hDM2 protein (as evidenced by HSQC 15N-1H NMR experiments). The peptidic foldamer also shows resistance to proteolytic degradation. |
|
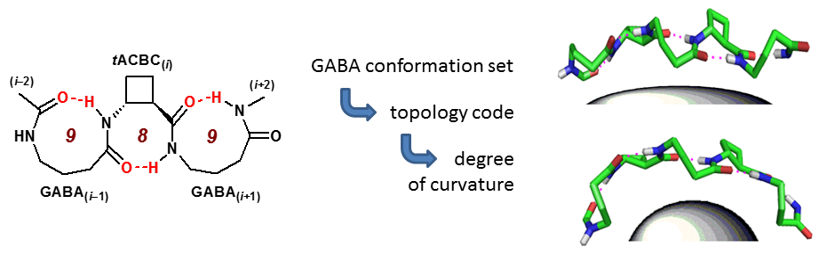
Using trans-ACBC and GABA as the monomer units to construct a mixed β,γ-peptide manifold, we discovered an unprecedented 9/8-ribbon, featuring an uninterrupted alternating C9/C8 hydrogen-bonding network. These ribbons adopt partially-curved topologies in which the ribbon is propagated incrementally along the peptide backbone either in a “straight” or “bent” manner, with the bend being in a plane perpendicular to that of the advancing ribbon’s backbone. We discovered a code which correlates the conformation of two successive GABA residues with the extent of ribbon curvature; intriguingly, the mathematical nature of this code makes it impossible to generate a 9/8-ribbon conformer having three or more consecutive “straight” or “bent” segments. This suggests that 9/8-ribbons cannot lie flat in a plane nor adopt highly-rounded structures: by benefiting from the continuous hydrogen-bonding network they are intrinsically destined to adopt partly-curved architectures. Chem. Commun., 2015, 51, 16233-16236.
|
Several low-energy conformers co-exist; they are more or less curved, but neither completely flat nor completely rounded. |
|
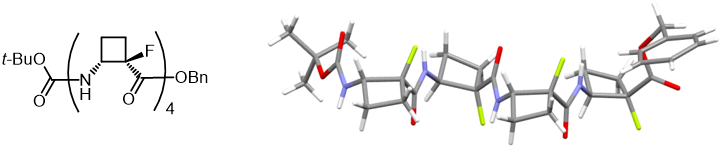
In contrast with peptides containing trans-ACBC, homo-oligomers of cis-ACBC adopt extended strand-like structures in which each residue appears to adopt a 6-membered hydrogen bond, as indicated above. When we examined the oligomers of α-fluorinated cis-ACBC, designed to destabilize the intra-residue 6-ring, these peptides retained the strand conformational preference, even though the implication of a 6-ring hydrogen bond was negligible. The backbone topology and the 4-membered ring orientations were perceptibly different from the non-fluorinated oligomers, with an inversion of the cyclobutane “butterfly” orientation. New J. Chem., 2015, 39, 3270-3279.
|
These oligomers adopt a regular zig-zag strand-like secondary structure which does not rely on intra- residue 6-ring hydrogen bonds for stability. |
|
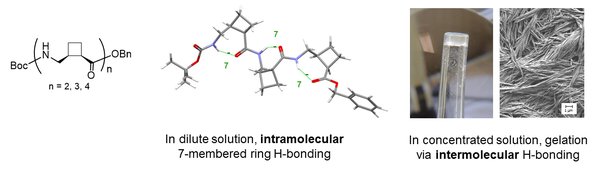
Homo-oligomers of cis-2-(aminomethyl)cyclobutane carboxylic acid, a cyclobutane γ-aminoacid display intriguing structural behavior, vacillating between inter- and intramolecular H-bonded conformations. In dilute solution, they adopt a number of low-energy conformers, including ribbon-like structures implicating a rare series of intramolecular 7-membered H-bonds. In concentrated solution, these interactions defer to an organized supramolecular assembly, leading to organogel formation and fibrillar xerogels. J. Org. Chem., 2017, 82, 4819-4828.